Dear list,
we have replicated gluster volumes much slower than the brick disks and I wonder if this is a configuration issue, a conceptual issue of our setup or really how slow gluster just is.
The setup:
- Three servers in a ring connected via IP over Infiniband, 100Gb/s on each link
- 2 x U.2 2TB-SSDs as RAID1, on a Megaraid controller, connected via NVMe links on each node
- A 3x replicated volume on a thin-pool LVM on the SSD-RAIDs.
- 2 x 26 cores, lots of RAM
- The volumes are locally mounted and used as shared disks for computation jobs (CPU and GPU) on the nodes.
- The LVM thinpool is shared with other volumes and with a cache pool for the hard disks.
The measurement:
- Iozone in automated mode, on the gluster volume (Set1) and on the mounted brick disk (Baseline)
- Compared with iozone_results_comparator.py
The issue:
- Smaller files are around factor 2 slower than larger files on the SSD, but factor 6-10 slower than larger files on the gluster volume (somewhat expected)
- Larger files are, except for certain read accesses, still factor 3-10 slower on the gluster volume than on the SSD RAID directly, depending on the operation.
- Operations with many, admittedly smaller files (checkouts, copying, rsync and unpacking) can extend into hours, where they take tens of seconds to few minutes on
disk.
- atop sometimes shows 9x% busy on some of the LVM block devices – and sometimes the avio value increases from 4-26ms to 3000-5000ms. Otherwise, there is nothing mentionable
observed regarding system load.
This does not seem to be network bound. Accessing from a remote VM via glusterfs-mount is not much slower despite connecting via 10GbE instead of 100GB. Nethogs shows sent traffic on the Infiniband
interface going up to 30000kB/s for large records – 240Mb/s over a 100Gb/s Interface ...
The machine itself is mostly idle – no iowaits, a load average of 6 on 104 logical cpus. Glusterfsd is sometimes pushing up to 10% CPU. I just do not see what the bottleneck should be.
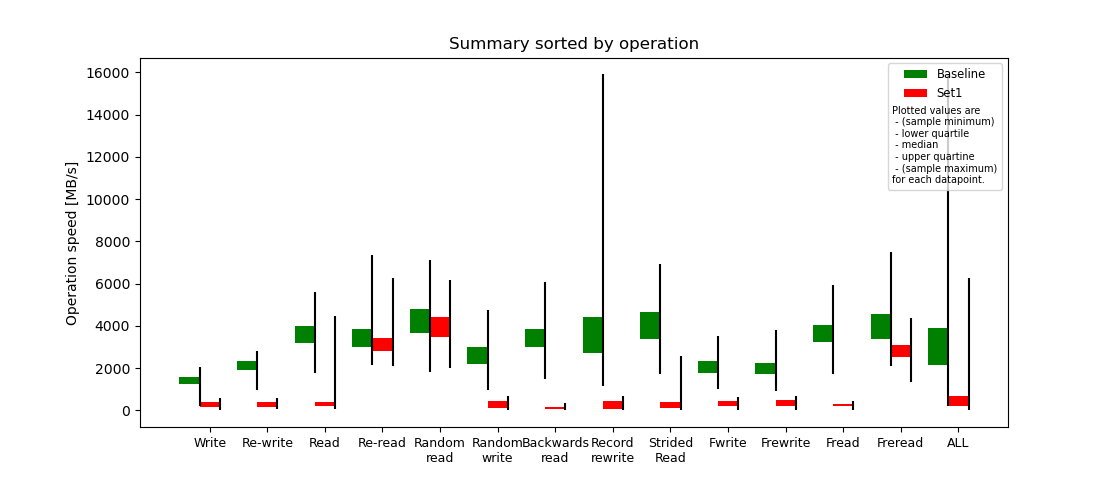
Below a summary out of the compared iozone reports.
Any hints if it is worth trying to optimize this at all and where to start looking? I believe I have checked all the “standard” hints, but might have missed something.
The alternative would be a single storage node connected via nfs, sacrificing the probably not needed redundancy/high availability of the replicated filesystem.
Thanks a lot for any comments in advance,
Best
Rupert
|
Operation
|
Write
|
Re-write
|
Read
|
Re-read
|
Random read
|
Random write
|
Backwards read
|
Record rewrite
|
Strided Read
|
Fwrite
|
Frewrite
|
Fread
|
Freread
|
ALL
|
|
baseline
|
first quartile
|
1260.16
|
1894.11
|
3171.44
|
3002.55
|
3679.12
|
2214.07
|
2987.15
|
2701.4
|
3361.62
|
1783.45
|
1739.6
|
3231.97
|
3405.8
|
2152.28
|
|
median
|
1474.31
|
2145.7
|
3637.6
|
3383.24
|
4167.16
|
2570.24
|
3435.44
|
3751.41
|
3967.55
|
2059.25
|
1992.64
|
3611.78
|
3936.96
|
3125.35
|
|
third quartile
|
1595.17
|
2318.0
|
3992.78
|
3840.0
|
4803.77
|
3013.13
|
3864.87
|
4420.33
|
4673.13
|
2354.6
|
2258.15
|
4036.73
|
4570.02
|
3915.54
|
|
minimum
|
194.64
|
960.92
|
1785.65
|
2152.15
|
1840.86
|
947.03
|
1491.1
|
1135.78
|
1732.88
|
999.01
|
933.54
|
1745.15
|
2083.72
|
194.64
|
|
maximum
|
2041.55
|
2819.84
|
5628.61
|
7342.68
|
7142.61
|
4776.16
|
6091.17
|
15908.02
|
6922.3
|
3540.22
|
3787.35
|
5933.0
|
7510.19
|
15908.02
|
|
mean val.
|
1408.31
|
2070.62
|
3663.03
|
3505.74
|
4231.31
|
2633.07
|
3444.43
|
4102.42
|
4029.73
|
2087.25
|
2027.23
|
3641.8
|
4113.02
|
3150.61
|
|
standard dev.
|
306.93
|
366.43
|
652.73
|
816.6
|
1048.38
|
781.84
|
796.13
|
2277.33
|
1041.12
|
511.69
|
464.33
|
687.27
|
1012.13
|
1335.36
|
|
ci. min. 90%
|
1363.0
|
2016.52
|
3566.67
|
3385.19
|
4076.54
|
2517.65
|
3326.9
|
3766.21
|
3876.03
|
2011.71
|
1958.68
|
3540.34
|
3963.6
|
3096.31
|
|
ci. max. 90%
|
1453.62
|
2124.71
|
3759.39
|
3626.3
|
4386.08
|
2748.5
|
3561.96
|
4438.62
|
4183.44
|
2162.79
|
2095.78
|
3743.27
|
4262.44
|
3204.91
|
|
geom. mean
|
1362.58
|
2034.2
|
3604.92
|
3422.21
|
4094.96
|
2511.93
|
3346.74
|
3637.05
|
3892.76
|
2024.11
|
1975.46
|
3576.11
|
3996.51
|
2883.44
|
|
set1
|
first quartile
|
173.4
|
174.43
|
208.58
|
2813.71
|
3498.77
|
139.63
|
51.51
|
91.49
|
124.34
|
201.55
|
199.05
|
187.72
|
2509.21
|
189.36
|
|
median
|
312.05
|
293.47
|
267.53
|
3134.22
|
4005.72
|
332.09
|
110.38
|
302.02
|
216.41
|
359.67
|
355.33
|
234.42
|
2742.35
|
357.31
|
|
third quartile
|
414.07
|
410.15
|
401.42
|
3446.69
|
4429.1
|
436.45
|
178.94
|
429.6
|
397.15
|
463.62
|
480.29
|
317.98
|
3092.73
|
686.35
|
|
minimum
|
44.11
|
45.6
|
59.07
|
2089.07
|
2004.07
|
9.39
|
8.67
|
2.77
|
32.58
|
38.63
|
41.09
|
26.0
|
1359.63
|
2.77
|
|
maximum
|
606.95
|
610.49
|
4457.8
|
6248.75
|
6153.85
|
666.55
|
374.16
|
696.91
|
2593.43
|
647.26
|
700.02
|
451.39
|
4353.18
|
6248.75
|
|
mean val.
|
308.37
|
309.17
|
544.27
|
3181.18
|
3964.96
|
305.76
|
123.49
|
282.89
|
402.01
|
337.11
|
342.11
|
248.76
|
2780.37
|
1010.03
|
|
standard dev.
|
154.66
|
157.37
|
892.64
|
635.03
|
825.98
|
183.62
|
87.03
|
191.43
|
493.51
|
160.15
|
173.1
|
100.31
|
484.33
|
1358.34
|
|
ci. min. 90%
|
285.54
|
285.93
|
412.49
|
3087.43
|
3843.02
|
278.65
|
110.65
|
254.63
|
329.15
|
313.47
|
316.56
|
233.95
|
2708.87
|
954.8
|
|
ci. max. 90%
|
331.2
|
332.4
|
676.05
|
3274.93
|
4086.9
|
332.87
|
136.34
|
311.15
|
474.86
|
360.76
|
367.67
|
263.57
|
2851.87
|
1065.27
|
|
geom. mean
|
260.27
|
260.16
|
318.88
|
3127.01
|
3872.62
|
220.54
|
86.71
|
174.2
|
243.43
|
285.24
|
286.02
|
223.44
|
2737.93
|
414.26
|
|
linear regression slope 90%
|
0.17 - 0.28
|
0.1 - 0.21
|
-0.05 - 0.35
|
0.83 - 0.97
|
0.85 - 1.0
|
0.08 - 0.15
|
0.02 - 0.05
|
0.04 - 0.07
|
0.04 - 0.17
|
0.12 - 0.2
|
0.13 - 0.22
|
0.05 - 0.09
|
0.6 - 0.72
|
0.28 - 0.35
|
|
ttest equality
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
DIFF
|
|
baseline set1 difference
|
-78.1 %
|
-85.07 %
|
-85.14 %
|
-9.26 %
|
-6.29 %
|
-88.39 %
|
-96.41 %
|
-93.1 %
|
-90.02 %
|
-83.85 %
|
-83.12 %
|
-93.17 %
|
-32.4 %
|
-67.94 %
|
|
ttest p-value
|
0.0
|
0.0
|
0.0
|
0.0005
|
0.026
|
0.0
|
0.0
|
0.0
|
0.0
|
0.0
|
0.0
|
0.0
|
0.0
|
0.0
|

